ggiraph
Make ggplot interactive
ggstance
Horizontal versions of ggplot2 geoms
ggalt
Extra coordinate systems, geoms & stats
ggforce
Accelarating ggplot2
ggrepel
Repel overlapping text labels
ggraph
Plot graph-like data structures
ggpmisc
Miscellaneous extensions to ggplot2
geomnet
Network visualizations in ggplot2
ggExtra
Marginal density plots or histograms
gganimate
Create easy animations with ggplot2
plotROC
Interactive ROC plots
ggthemes
ggplot themes and scales
ggspectra
Extensions for radiation spectra
ggnetwork
Geoms to plot networks with ggplot2
ggtech
ggplot2 tech themes, scales, and geoms
ggradar
radar charts with ggplot2
ggTimeSeries
Time series visualisations
ggtree
A phylogenetic tree viewer
ggseas
Seasonal adjustment on the fly
ggtree
https://bioconductor.org/packages/release/bioc/html/ggtree.html
gtree is designed for visualizing phylogenetic tree and different types of associated annotation data.
# Example from https://bioconductor.org/packages/release/bioc/html/ggtree.html
library(ggplot2)
library(ape)
library(ggtree)Tree Data Import
file <- system.file("extdata/BEAST", "beast_mcc.tree", package="ggtree")
beast <- read.beast(file)Users can use ggtree(beast) to visualize the tree and add layer to annotate it.
ggtree(beast, ndigits=2, branch.length = 'none') + geom_text(aes(x=branch, label=length_0.95_HPD), vjust=-.5, color='firebrick')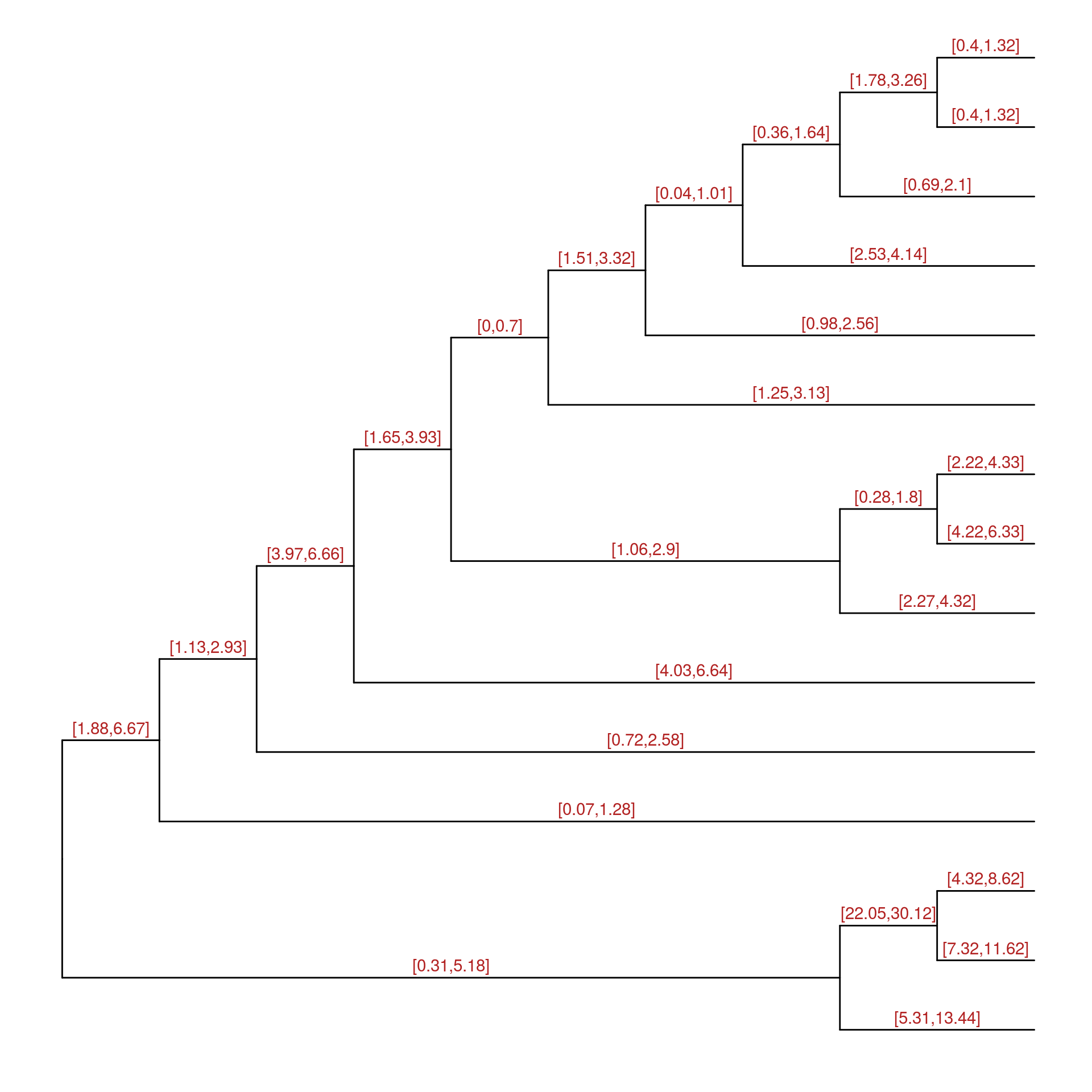
Tree Visualization
To view a phylogenetic tree, we first need to parse the tree file into R. The ggtree package supports many file format including output files of commonly used software packages in evolutionary biology. For more details, plase refer to the Tree Data Import vignette.
library("ggtree")
nwk <- system.file("extdata", "sample.nwk", package="ggtree")
tree <- read.tree(nwk)
ggplot(tree, aes(x, y)) + geom_tree() + theme_tree()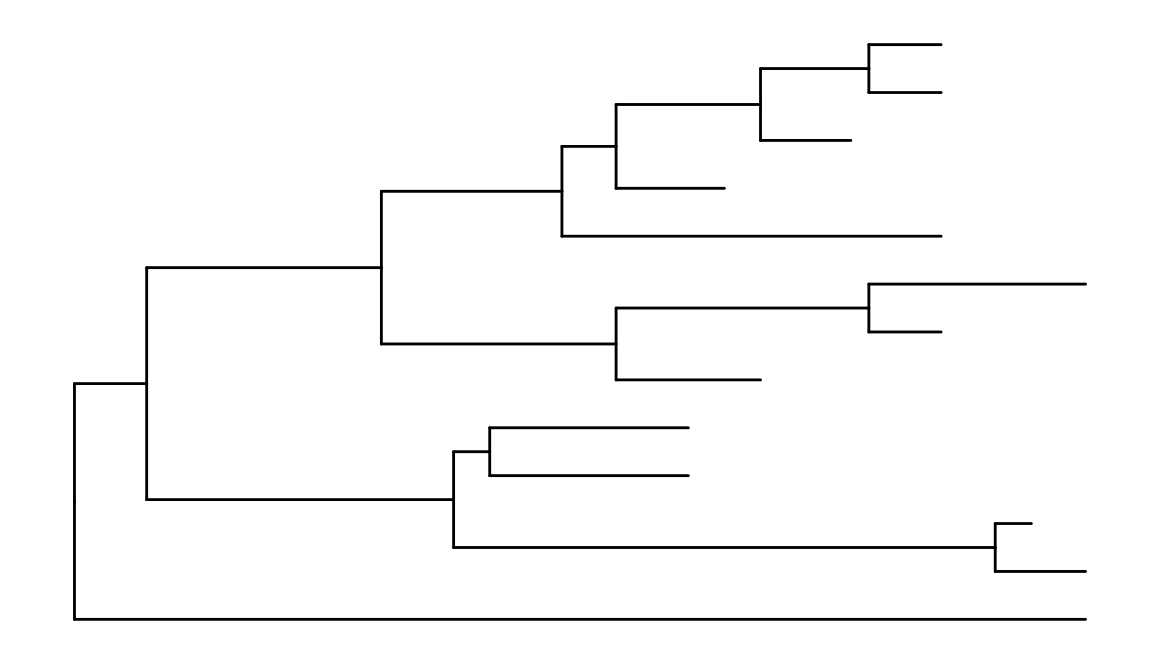
Tree Manipulation
Internal node number
Some of the functions in ggtree works with clade and accepts a parameter of internal node number. To get the internal node number, user can use geom_text2 to display it:
nwk <- system.file("extdata", "sample.nwk", package="ggtree")
tree <- read.tree(nwk)
ggtree(tree) + geom_text2(aes(subset=!isTip, label=node), hjust=-.3) + geom_tiplab()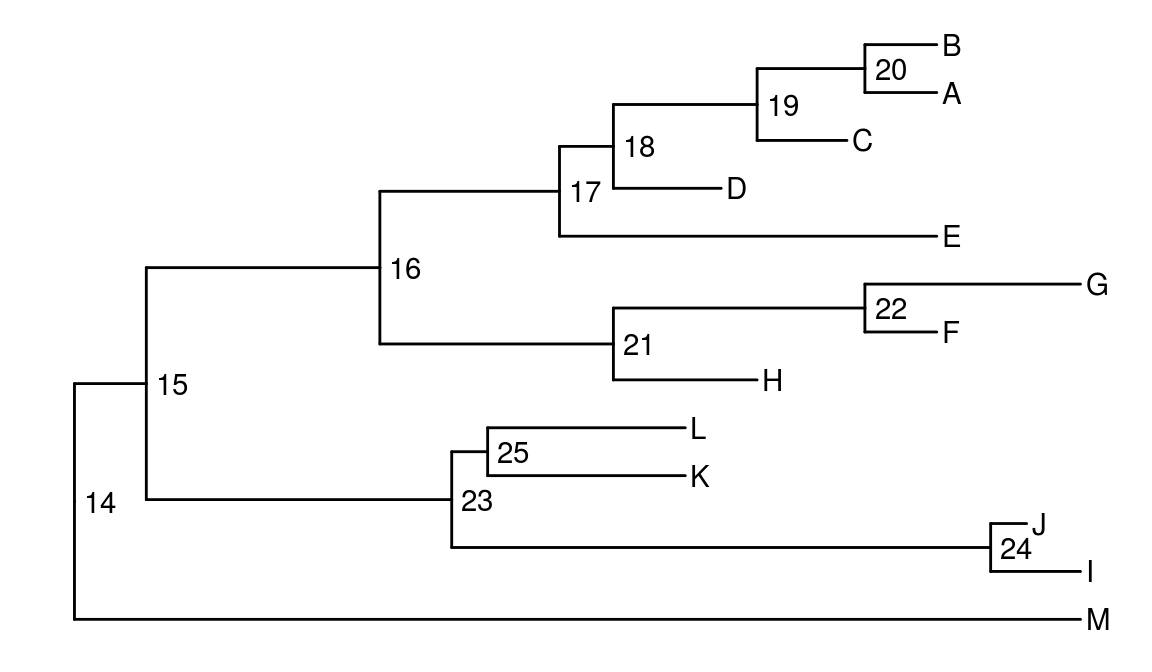
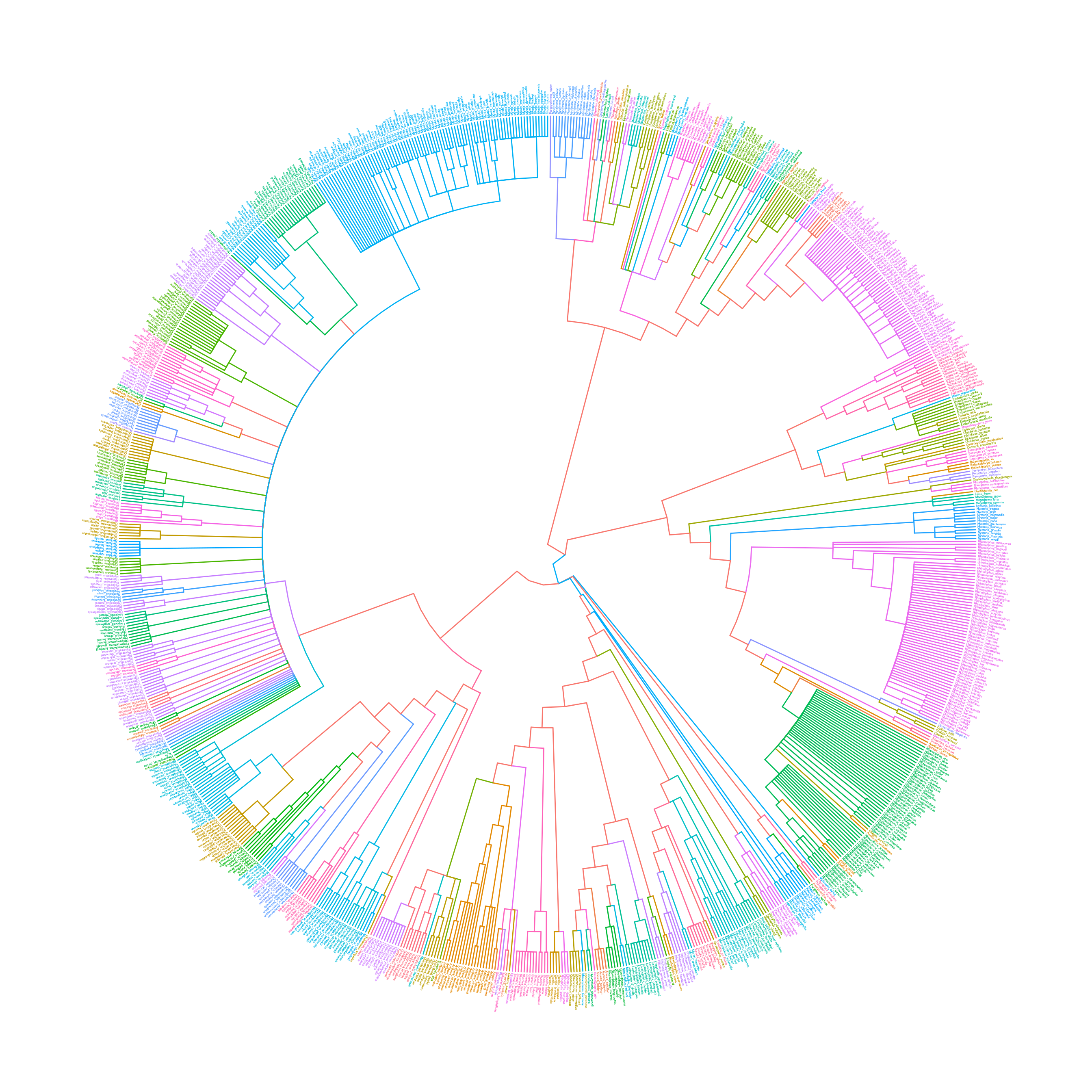
The following example use groupOTU to display taxa classification.
data(chiroptera)
groupInfo <- split(chiroptera$tip.label, gsub("_\\w+", "", chiroptera$tip.label))
chiroptera <- groupOTU(chiroptera, groupInfo)
ggtree(chiroptera, aes(color=group), layout='circular') + geom_tiplab(size=1, aes(angle=angle))